This is meant to be a brief look at
Proteome Cluster.
We just wanted to
see how easy it was to get started and use. If you would like to do a similar demo you can request a
temporary password from Bioproximity.
We went into this demo naive just to see what we would
run into. We feel that naive tests are the best tests. The first thing we found out
was that the program is not
compatible with Microsoft's Internet Explorer. (Since we wrote that
last sentence PC has fixed this problem and it has been tested on IE9.
They are responsive, hmm we are not used to that.) Previously, we switched over to
the most current version of FireFox and everything worked. Other
compatible browsers and file formats are listed on their website.
If you would like to repeat the demo for yourself go to
the bottom of this page and download the data files that we used, or use
your own data file.
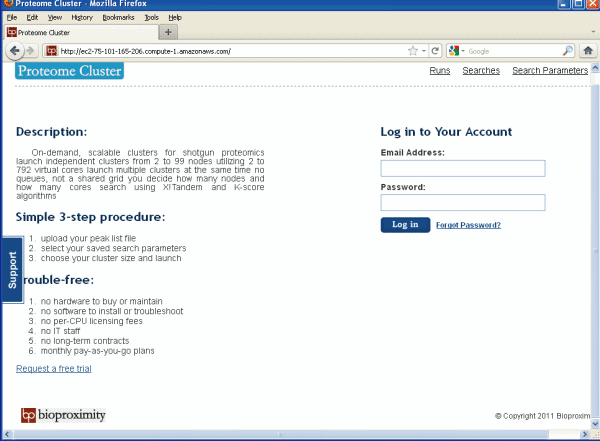
This is what the log in page looks like. It is very simple, see Figure 1 below.
Figure 1.
Log In Page
As shown in Figure 2 below there are really only
four pages to explore: Upload, Runs, Searches and Search Parameters.
Figure 2.
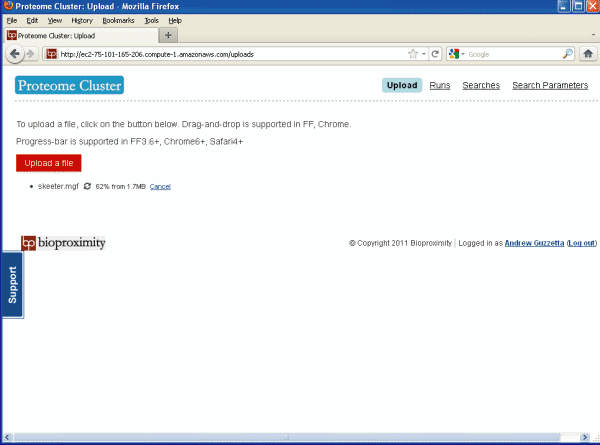
The upload page as shown in Figure 2, above, is where you
upload your files to be searched. Just press on the "Upload a file"
button and browse to a file on your hard drive. The compatible file
formats are MGF, mzML or
mzXML. We will be uploading our data in MGF format, "Mascot Generic Format".
The MGF format was developed
by Matrix-Science,
http://www.matrixscience.com/help/data_file_help.html At
first we tried to upload concatenated DTA files (an old Thermo data
format), but that is not an
acceptable file format for Proteome Cluster, as of yet. If you have
individual DTA's concatenate them into MGF format with the tools
provided by Matrix-Science,
http://www.matrixscience.com/downloads/merge.zip . Once
these tools are downloaded and unzipped use the merge.pl tool to make your MGF file.
Many MS manufacturers will allow you to export your proteomics data
in the allowable formats without the need for conversion.
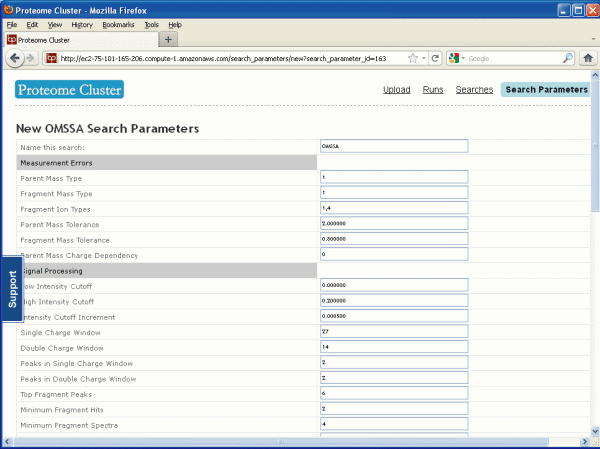
As usual before you can start a search you must
decide on your search parameters. We decided to do an OMSSA
search. Here is what the search parameters page looks like, see
Figure 3 below. We will talk more about OMSSA later. For the most part the
default parameters will work, but you may want to change the mass
tolerances depending on the capability of your instrument and you
will want to choose a species database that is appropriate for your
sample. Proteome Cluster currently supports four search
algorithms: OMSSA, X!Tandem, X!Tandem using k-score (also called
Comet), and X!Hunter.
Figure 3
Search Parameters Page
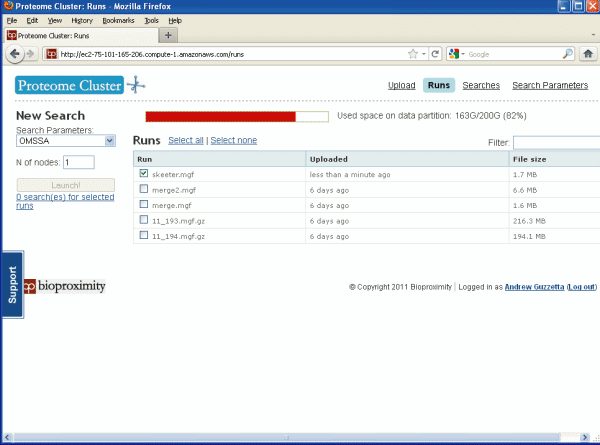
The "Runs Page" below lets you select which file should be
searched and also allows you to start the search. Just check
the box next to your uploaded data file and click "Launch!" It
will ask you to confirm the search, this is a safety feature since outside of the demo
the search
time will cost you money.
Figure 4.
Runs Page
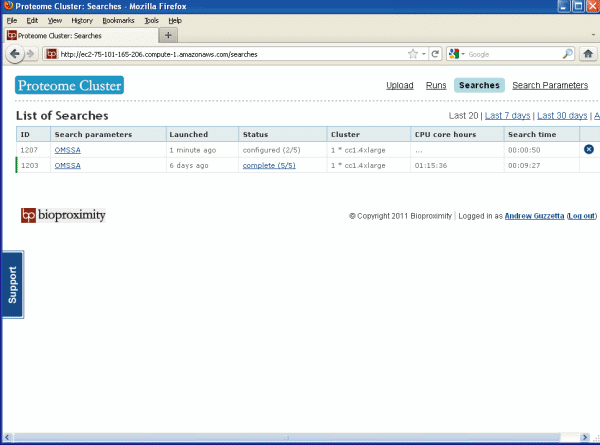
Go to the Searches page, see Figure 5 below, to see
if your search is done. Once it says "complete" you can
download your OMSSA results. Just click on the link to
download.
Figure 5.
Searches Page
To view your OMSSA proteomic result you will need to
download OMSSA and choose to open the results file that you just
downloaded. You can download the OMSSA browser free at
http://pubchem.ncbi.nlm.nih.gov/omssa/browser.htm. You can also
do searches on your local computer using OMSSA. To learn more
about OMSSA we have a review here,
http://www.ionsource.com/functional_reviews/OMSSA/OMSSA_Rev.htm
We did not review the X!Tandem search.
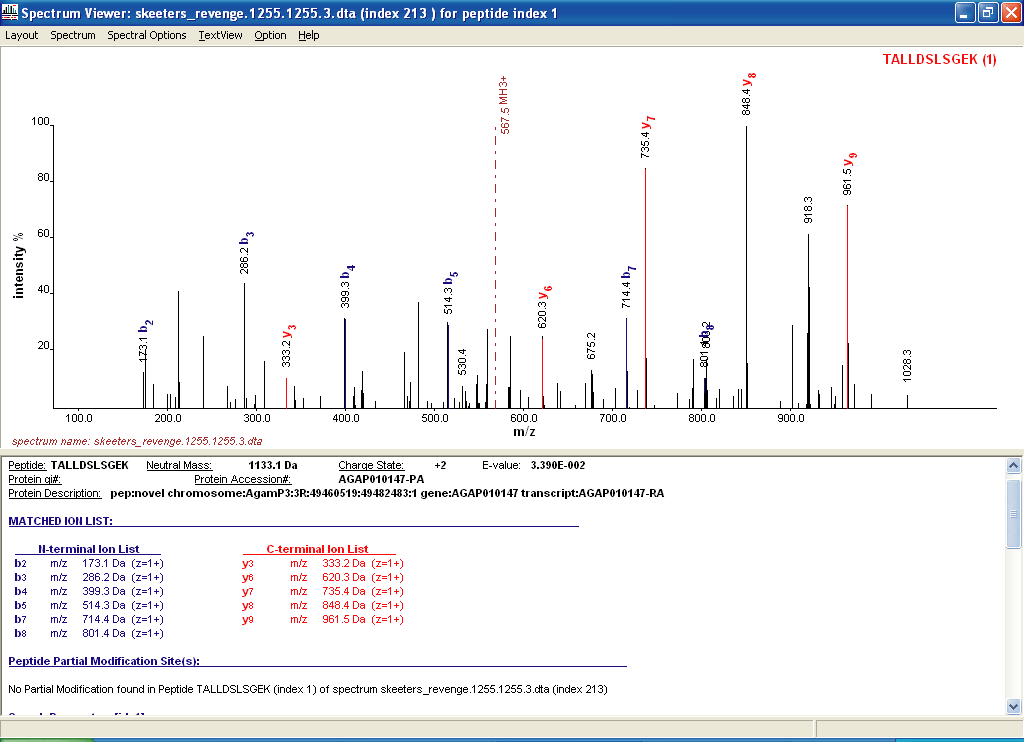
Figures 6 and 7 below shows you some of the things
that you can see with your OMSSA browser. In Figure 6 you can
see the protein coverage of one of the hits, and in Figure 7 you can
see one of the MS/MS matches of one of the peptides.
Figure 6.
OMSSA Protein Coverage Details
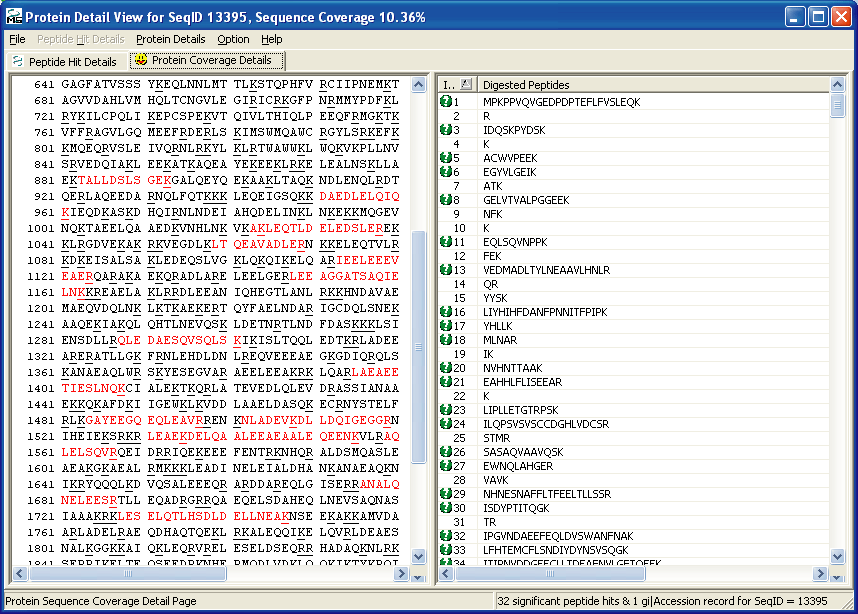
Figure 7.
OMSSA MS/MS spectral match
Conclusion: Proteome Cluster was easy to use
and the web interface was intuitive. We did not go into an
exhaustive review of the product, and only wanted to take a look,
and see what it was, and how easy it was to use. Proteome Cluster
was simple, and we know that simple and intuitive isn't always so
simple to produce, so good job Proteome Cluster, and Bioproximity.
We did run into a few problems but that is because we did not read
the instructions right in front of us. We think that this is a real
fine place for Proteome Cluster and Bioproximity to start. We
will be checking back soon.
Links:
1.) Bioproximity
www.bioproximity.com
2.) Proteome Cluster www.proteomecluster.com
3.) Watch a YouTube
video of Proteome Cluster
4.) Request a Proteome Cluster
demo
Extras:
1.) Download the Xcalibur (ThermoFisher)
raw data file that we used in this
tutorial. (zipped 9.5 MB)
2.) Download the zipped dta
files associated with the above data file (zipped 3.3 MB)
3.) Visit Bioproximity.com to get a temp demo
password.
4.) Download the concatenation tools from
Matrix-Science if you need them.
You can use these files courtesy of IonSource for your own fun or comparisons,
but not for profit. Feel free to to use them in any non-profit publication
or presentation you wish as long as you cite IonSource.com
as the source of the data. We paid about $350 for that data
file.
e-mail the webmaster@ionsource.com
with all inquiries
home
| terms of use
(disclaimer)
Copyright © 2011-2016 IonSource, LLC All rights reserved.
Last updated:
Tuesday, January 19, 2016 02:51:47 PM
|







