|
|
||
|
|
||
|
De Novo Peptide Sequencing Tutorial Example Spectrum part one, low mass |
||
|
|
||
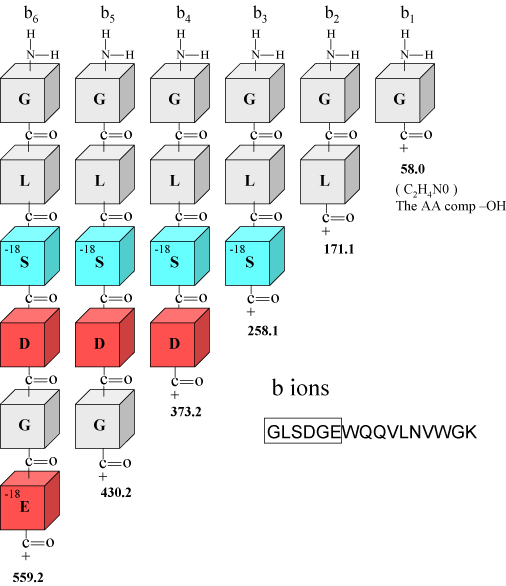
| For this example we will start at the low end of the spectrum and work our way up to the high end. This is just an example spectrum used for illustrative purposes. We will basically know the ions we are looking for and go look for them, and also we will learn what we can learn. Later on with the exercises we will follow the rules of our protocol and start at the high end of the spectrum. Below, in Figure 1, are six of the b ions described previously for the tryptic peptide GLSDGEWQQVLNVWGK. You may notice that we have annotated the serine and glutamic acid blocks with the notation -18, this is to remind us that these residues may be associated in the spectrum with water losses. In Figure 2 we have made a screen shot of the low end of the MS/MS spectrum to look for some of the predicted b ions and their associated fragment ions. | ||
|
b Ion Details
Figure 1. |
||
|
|
||
|
Figure 1. We have taken the first 6 amino acids from the peptide GLSDGEWQQVLNVWGK to show in detail, b ions 1-6. The calculated b ion masses are noted in bold numbers below the block figures. |
||
|
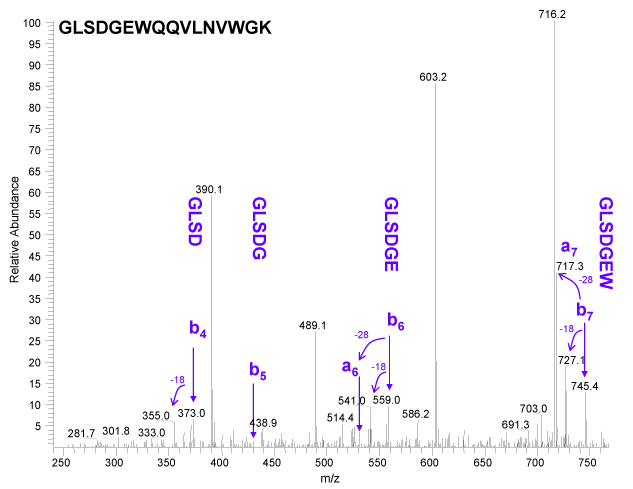
In Figure 2 below, it is easy to see the that the b ion series is not the predominant series. Data was not acquired below m/z 240 because this is ion trap data, and traps typically have a low mass cutoff, sometimes called the 1/3 rule. Due to this fact we have lost our diagnostic low mass immonium ions and several other important diagnostic ions. We were easily able to observe b ions 4, 6 and 7. Ion b5 was too low to call, most likely due to to the overall low abundance of the b ion series at the lower end of this spectrum. Calling a true de novo sequence with this b ion data would have been a challenge with these low ion intensities. There is an interesting feature in this spectrum that confirms that we are looking at a b ion series, which is that we observe a ions associated with b6 and b7. The loss of C=O or 28 u is the characteristic mass difference for the a / b pair. One can also observe water losses for b ions 4, 6 and 7 as predicted by the presence of serine and glutamic acid in this sequence.
Figure 2.
|
||
| Why did the b5 ion drop out in Figure 2? It looks like there was a significant water loss with a peak at m/z 412 which could have diminished the already small peak at m/z 430? Or, it could be the de novo sequencing rule, "b ion intensity may drop when the next residue is P, G or also H, K, and R", pretty cool, huh! |
||
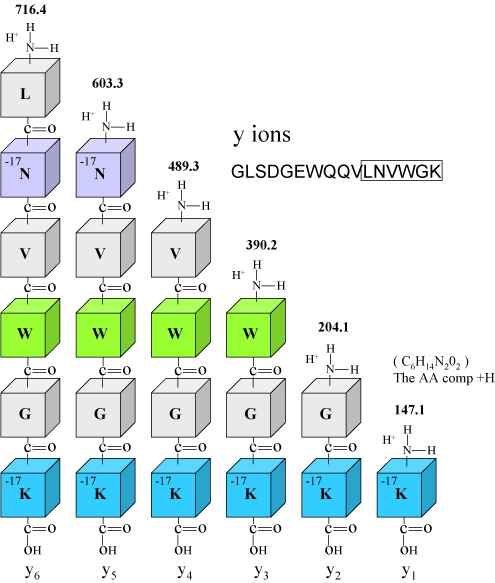
| Figure 3.
|
||
|
|
||
|
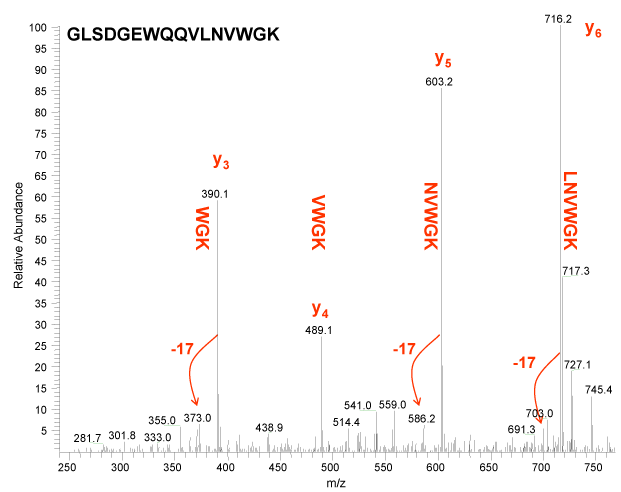
Figure 3 above illustrates the y ion series. Again note that we have annotated the lysine and asparagine blocks with -17. This is to remind us that these residues appear to lose ammonia, and for the majority of y ions observed below in Figure 4, we also observe a peak that is 17 u lower. The apparent loss of ammonia is diagnostic for these residues.
|
||
|
Figure 4.
|
||
|
So far we have discussed b and y ions, and for simplicity we have relegated
ourselves to the terminal 6 or 7 amino acids. We will detail the
entire spectrum in the following slides. It is apparent that the
amino terminal sequence may be difficult to determine from the b ion series,
one because of the relative abundance, and two because of the limitations of
the ion trap. At the end of this tutorial we will provide spectra
obtained on a q-TOF mass spectrometer for you to practice with, which does
not share the same ion trap low mass limitation.
If you are interested in following this sequence proceed onto the next page. It is getting kind of interesting, isn't it? I wonder what will happen next? |
||
| - | ||
|
|
||

|
||
| back | forward | |
|
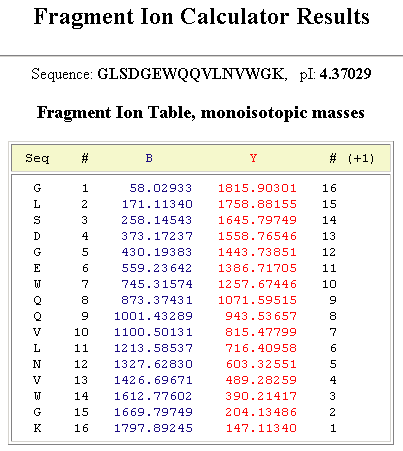
Reference Figuresb and y Ion Table
|
||
|
|
||
|
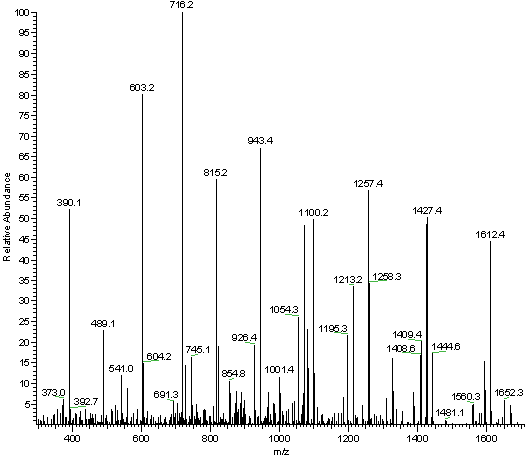
Full Fragment Spectrum
|
||
|
|
||
|
|
||
|
|
||
| e-mail the webmaster@ionsource.com with all inquiries | ||
| home
| terms of use
(disclaimer) Copyright � 2012 IonSource All rights reserved. Last updated: Monday, February 01, 2016 11:04:41 AM |
||
|
|
||